Supplies
Sylgard™ 184 silicone elastomer package (known as polydimethyl siloxane or PDMS) was obtained from Dow Silicones Company (Midland, MI). Polyester (PETE) membrane filters (clear, 0.2 µm pore dimension, 12 µm thickness, 2e6 pores/cm2) have been from Sterlitech Company (Kent, WA). Pre-cleaned 75 mm × 50 mm × 1 mm plain glass slides have been from Corning Included (Corning, NY). INTRAMEDIC polyethylene (PE) tubing (I.D. 1.40 mm, O.D. 1.90 mm; and I.D. 1.14 mm, O.D. 1.57 mm) have been from Clay Adams. Silicon (SI) wafers have been from College Wafer Inc (Boston, MA). SU-8 2025 photoresist and SU-8 developer (98–100% 1-Methoxy-2-propyl acetate) have been from Kayaku Superior Supplies Inc (Westborough, MA). MIL PRF 131k Class 1 foil, 150 µm thick (MarvelSeal® 470) was from Berry World (Evansville, IN). A1 body, a skinny polymer movie (A4 inkjet waterproof movie) was from CisInks (South El Monte, CA). Polyethylene glycol diacrylate (PEGDA; 5 kDA), 4-arm poly(ethylene glycol)-acrylate (4-arm PEG-Ac; 10 kDa), and poly(ethylene glycol)-dithiol (PEG-diSH; 3.4 kDa) have been from Laysan Bio Inc. (Arab, AL). Phosphate buffered saline (PBS, 10 X, pH 7.4), Trypan Blue (0.4%), fluorescent polystyrene beads (Fluoro-Max™ Fluorescent Pink, 542/612 nm, d = 2 µm), and Temozolomide (TMZ) have been from Thermo Fisher Scientific (Waltham, MA). Irgacure 2959 was from BASF company (Florham Park, NJ). Fetal bovine serum (FBS) and penicillin/streptomycin (P/S) have been from Hyclone (Logan, UT). Roswell Park Memorial Institute (RPMI)-1640 medium and 0.05% trypsin/0.02% ethylenedinitrilotetraacetic acid (EDTA) have been from Coring (Coring, NY). Acridine orange (AO) and carmustine (BCNU) have been from Millipore Sigma (St Louis, MO). Propidium iodide (PI) was from MP Biomedical LLC (Solon, OH). Cell stain 3,3’-dioctafecyloxacarbocyanine perchlorate (DiOC) was from Life Applied sciences (Carlsbad, CA). Good Blue FCF (CAS Quantity: 3844-45-9) was bought as blue meals coloring from an area grocery retailer. Homo sapiens mind glioblastoma U87 cells have been from ATCC (Manassas, VA). Glycine–Arginine–Cysteine–Aspartic Acid–Arginine–Glycine–Aspartic Acid–Serine (GRCD-RGDS), Glycine–Arginine–Cysteine–Aspartic Acid–Arginine–Glycine–Aspartic Acid–Serine–FITC (GRCD-RGDS-FITC), and Aspartic Acid–Arginine–Cysteine–Glycine–Valine–Proline–Methionine–Serine–Methionine–Arginine–Glycine–Cysteine–Arginine–Aspartic Acid (DRCG-VPMSMR-GCRD) peptides have been from Genic Bio (Shanghai, China).
Microfluidic machine preparation
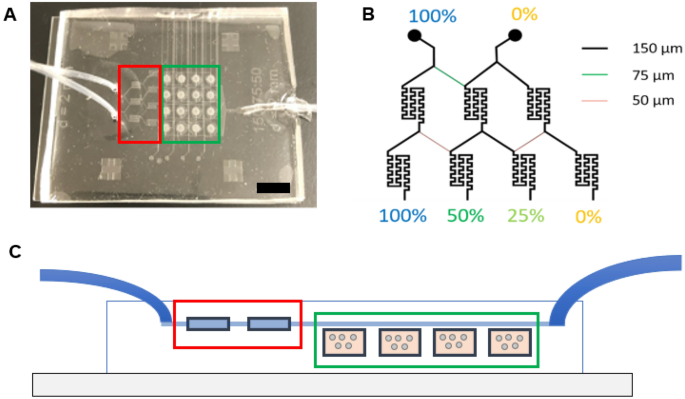
The microfluidic machine was fabricated by assembling a help glass microscope slide (75 mm × 50 mm × 1 mm), two layers of patterned PDMS (prime perfusion layer and a backside cell tradition chambers layer with cell loading channels), sixteen PETE membranes (capping the sixteen cell tradition microwells), and three tubes—two inlet tubes (PE tubing; I.D. 1.14 mm, O.D. 1.57 mm) and one outlet tube (PE tubing; I.D. 1.40 mm, O.D. 1.90 mm) (Fig. 1, Supplemental Fig. S1). The highest perfusion layer was 3 mm thick and the underside cell tradition microwells layer was 250 µm thick. The peak of the perfusion and cell loading channels was 50 µm. A nicely depth of 250 µm was chosen to be bigger than a person cell whereas nonetheless permitting imaging via the whole pattern. Most perfusion channels have been 150 µm extensive with the important thing exceptions of fifty µm and 75 µm, which have been integrated into the design to realize fractional dilution sequence. These fractional dilution channels have been used to supply a focus gradient of 100%, 50%, 25%, and 0% of preliminary solute focus. The cell loading channels on the underside layer have been additionally 150 µm extensive.
Microfluidic machine design. (A) Picture of the microfluidic machine with the inlet (left-hand facet of picture) and outlet (right-hand facet of picture) tubing. Black scale bar represents 1 cm. (B) Channel widths of the microchannels and the focus dilutions generated with these channel widths. (C) Facet view of the fabricated microfluidic gadgets displaying the inlet tubes (left), mixing chambers (pink), cell tradition chambers (inexperienced), and outlet tube (proper).
Each prime and backside PDMS layers have been fabricated through curing PDMS precursor on patterned Si masters (fabricated by customary photolithography SU-8 2025 process: https://kayakuam.com/wp-content/uploads/2019/09/SU-82000DataSheet2025thru2075Ver4-3.pdf) at 75 °C in a single day. Briefly, a 50 µm thick SU-8 2025 photoresist was spin-coated on a Si wafer at 1700 rpm (SP-100 spinner, BidTec) and tender baked on a hotplate at 65 °C for 3 min after which at 95 °C for 9 min. The patterns on the pre-printed masks have been transferred to the photoresist layer via UV radiation (160 mJ/cm2 with Flood Publicity Mannequin 60, ABM-USA, Inc., San Josa, California, USA), post-exposure baked (on a hotplate at 65 °C for six min after which at 95 °C for 7 min) and developed by an ordinary course of proven in Supplemental Fig. S2. UV radiation crosslinked the uncovered SU-8, and SU-8 developer dissolved the uncrosslinked SU-8, leaving solely the crosslinked SU-8 patterns.
The underside cell tradition PDMS layer was ready equally with a modification to regulate thickness (Supplemental Fig. S3). To attain a thickness of 250 µm for the underside PDMS layer, a 150 µm Al foil body spacer was used. After pouring PDMS precursor on the Si grasp and the Al body, a skinny polymer movie and a inflexible glass slide have been placed on prime. The versatile polymer movie was added to keep away from the contact of PDMS precursor and the glass slide, in any other case the inflexible glass slide couldn’t be eliminated after PDMS was cured. A 5 lb. weight was added on the highest to make sure a uniform thickness of the PDMS layer, and the PDMS was cured at 80 °C in a single day. The thickness was measured by an Alpha-Step IQ profilometer (KLA-TENCOR Alpha D500, KLA Corp., Milpitas, California, USA). Be aware that the PDMS layer was thicker than the spacer as not all extra PDMS was extruded via the perimeters previous to polymerization because of the excessive viscosity of the PDMS precursor.
After the PDMS layers have been cured, 16 cell chambers (2 mm I.D.), 2 inlets (1.5 mm I.D.) and 1 outlet (2 mm I.D.) have been lower with biopsy punches. Sixteen round PETE membranes (pore dimension 0.2 µm), lower with 3 mm diameter biopsy punch, have been individually positioned over every of the two mm ID cell tradition microwells to separate the 2 PDMS layers and forestall cells and materials within the tradition chambers from escaping into the perfusion channels. Particular person membrane items have been chosen versus one piece to allow higher adhesion between the 2 PDMS layers (prime and backside layers of the machine), to keep away from leakage, and to guarantee stability over time. Particular person items additionally forestall the crosstalk between wells, which might occur with one piece membrane layer. The three mm diameter membranes are barely bigger than the two mm ID wells, so the perimeters of the membranes contact the sleek surfaces of the PDMS, adhere to it spontaneously, and keep regular throughout the next oxygen plasma and alignment. A laboratory corona therapy machine (BD-20AC mannequin, Electro-Technic Merchandise, Chicago, IL, USA) was then used to deal with the 2 PDMS surfaces with plasma for 1 min to oxidize them. The 2 PDMS layers have been aligned with 4 registration marks on the corners by eye. After the 2 PDMS layers have been pressed along with PETE membranes in-between, the glass slide surfaces and PDMS meeting have been additional oxidized with plasma for 1 min to enhance attachment. Inlets and outlet tubing have been inserted into the inlets and outlet places and sealed with uncured PDMS. The gadgets have been heated at 75 °C for at the least 2 h to enhance sealing and treatment the PDMS. An in depth machine meeting schematic is proven in Supplemental Fig. S4.
Mixer efficiency and dilution evaluation
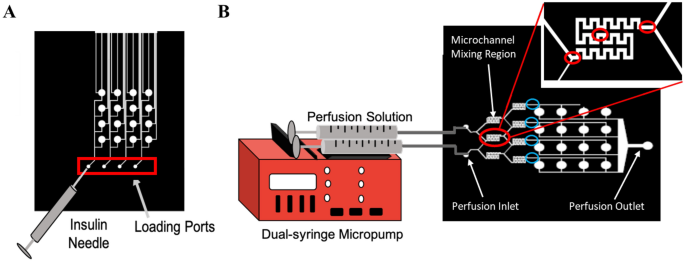
For perfusion, perfusion fluids have been positioned in 3 mL syringes in a twin syringe pump (New Period Pump Programs Inc, Farmingdale, NY) so the inputs into each side of the mixers had the identical movement charge. The syringes have been related to the inlet ports of the machine with needles (21 G) and tubing (PE tubing I.D. 1.14 mm, O.D. 1.57 mm). Volumetric movement charges examined have been 1, 1.5, 2, 5, and 10 µL/min. To check mixing effectivity, an answer of Good Blue FCF (to help in visualization) was dissolved in PBS for a remaining focus of 25 μM (measured through Beer-Lambert Legislation)23 and utilized in mixture with plain PBS to find out the efficiency of the micromixers. Utilizing an inverted microscope (Zeiss, Axiovert 200 M, Oberkochen Germany) at 5X magnification, photos have been captured at three areas—entrance, center, and exit—of the micromixers throughout perfusion (Fig. 2B). ImageJ software program24 was then used to separate out RGB channels and measure the depth of the blue RGB channel throughout the micromixers (as a perform of micromixer width) utilizing the ‘Plot Profile’ perform. Values have been normalized from 0–1 for every set of photos, with a grey worth of 1 being assigned to the blue shade, and a grey worth of 0 assigned to the background shade of the microfluidic chip. Absolutely the mixing index (AMI) was calculated as:
$$AMI = frac{sigma }{< I >}=frac{sqrt{frac{1}{N}{sum }_{i=1}^{N}({I}_{i}- {< I >}^{2}}}{< I >},$$
(1)
the place Ii is the native pixel depth, < I > is the imply pixel depth within the cross part, and N I the full variety of pixels25.
Loading and perfusion set-up. (A) Schematic of loading the hydrogel precursor resolution into the membrane-capped microwells of the underside loading layer of the machine through the loading ports utilizing an insulin needle. (B) Schematic of perfusion set-up by which tubing connects the three mL syringes containing the perfusion fluids to the inlet ports of the microfluidic machine. Pink circles point out the place photos have been taken throughout the micromixer for mixing evaluation (entrance, center, and exit). Blue circles point out the place photos have been taken on the finish of the micromixer for focus gradient evaluation.
As well as, to find out the focus gradient generated by the microfluidic machine on the set volumetric movement charges, photos have been additionally captured on the finish of every micromixer and ImageJ software program was then used to check the common depth of the dye produced by the micromixers.
Calculation of diffusion coefficient, hydrodynamic radius, and diffusion time of molecules via the width of the blending channel
Modeling molecules as laborious spheres, we used the Stokes–Einstein equation to calculate the diffusion coefficient for molecules:
$$D=frac{kT}{6pi mu r},$$
(2)
the place D is the diffusion coefficient, (ok) is the Boltzmann fixed, (mu) is the dynamic viscosity of water at 37 °C, and (r) is the hydrodynamic radius of the particle. Assuming that the molecular quantity of the particle corresponds to a spherical form, the hydrodynamic radius of every molecule was calculated as:
$$r=sqrt[3]{frac{3frac{MW}{{N}_{A}rho }}{4pi }},$$
(3)
the place MW is the molecular weight of the particle (supplied by the Royal Society of Chemistry), NA is Avogadro’s Quantity, and (rho) is the density of the particle (supplied by the Royal Society of Chemistry). The time required for a molecule to diffuse throughout the width of the channel was then calculated as:
$$t=frac{{x}^{2}}{2D},$$
(4)
the place (t) is the time required for diffusion to happen and (x) is the diffusion distance.
Cell upkeep
Human glioblastoma U87 cells have been cultured in RPMI-1640 medium supplemented with 10% FBS and 1% P/S and incubated in a humidified incubator at 37 °C and 5% CO2. Cells have been passaged by a 5 min publicity to Trypsin/EDTA as soon as an ~ 80% confluency was achieved, and media was modified each 2 days. Cell passages 10–20 have been used for experiments. Cells have been cultured with DiOC (20 μM) for twenty-four h to stain all cells previous to all additional experimentation.
Loading of PEG hydrogels within the cell tradition chambers of the microfluidic machine
Non-degradable, non-adhesive PEGDA hydrogels have been ready by UV photo-polymerization. Briefly, to organize a inventory resolution of the photoinitiator, 1% w/v Irgacure 2959 was dissolved in de-ionized water (DI water), sonicated (Branson, Mannequin #2800, 40 kHz) for 90 min and saved at room temperature, protected against the sunshine for as much as 2 weeks. PEGDA hydrogel precursor resolution (250 µL) of 20% w/v and 0.1% w/v in Irgacure have been ready in PBS and vortexed for 30 s to guarantee full mixing. The hydrogel precursor resolution was loaded into the cell tradition microwells of the microfluidic machine via the loading ports through the use of an insulin needle (Fig. 2A). Every microwell contained roughly 0.2 µL of the hydrogel precursor resolution; therefore 3.14 µL of resolution have been wanted to fill all 16 of the microwells. The surplus quantity of precursor resolution was essential to make sure that the answer reached the microwells as an alternative of remaining throughout the loading channels. Aid channels have been integrated into the design of the loading layer of the machine to allow the surplus precursor resolution to exit the machine as an alternative of pushing in opposition to the membrane capping the microwells. The hydrogels have been then polymerized beneath UV lamp (4.81 mW cm2, 1 W, 365 nm; Blak-Ray® XX-15L UV bench lamp, UVP, Upland, CA) for 10 min26. These gels are known as PEGDA all through.
Degradable, adhesive 4-arm PEG-Ac hydrogels have been ready by Michael-type addition. First, 4-arm PEG-RGDS was ready following a beforehand developed protocol27. We modified on common one of many acrylate teams of every 4-arm PEG-Ac with RGDS (80% modification effectivity) and saved the lyophilized product beneath argon in a desiccated container at − 20 °C for as much as 6 months. Hydrogels have been then fashioned utilizing a combination of 4-arm PEG-Ac and 4-arm PEG-RGDS (0.8 mM in RGDS) and a 50:50 crosslinker combination of PEG-diSH and an enzymatically degradable peptide crosslinker DRCG-VPMS↑MR-GCRD. For all gels, 20% w/v inventory options of 4-arm PEG-Ac, PEG-diSH, and 4-arm PEG-RGDS have been ready in TEA buffer pH 8 instantly prior to make use of. Inventory options have been combined and the peptide crosslinker was added as powder to present a hydrogel precursor resolution with a 1:1 acrylate to thiol molar ratio and a remaining polymer focus of 10% w/v and a remaining peptide crosslinker focus of seven.8 mM. The hydrogel precursor resolution was combined through pipetting for 30 s, injected within the microfluidic machine as described above, and allowed to gel for 20 min in a humidified incubator at 37 °C and 5% CO2. These gels are known as 4-arm PEG-Ac all through.
To evaluate the reproducibility of hydrogel and cell loading into the cell tradition microwells of the microfluidic machine, DiOC-stained U87 cells have been added to the PEGDA hydrogel precursor resolution at 106 cells/mL. The precursor resolution was then loaded into the microwells utilizing an insulin needle and gelled beneath UV as described above. Photos of every nicely have been taken utilizing an inverted fluorescence microscope and ImageJ was used to depend the variety of ells inside every nicely. The distribution of the fluorescently labelled cells throughout the hydrogel was imaged utilizing a confocal microscope (Leica Confocal SP8, Leica Microsystems Inc, Buffalo Grove, IL) at × 10 magnification and processed utilizing Fiji software program (free obtain http://fiji.sc). For confocal imaging the hydrogel was additionally rendered fluorescent by covalently incorporating a fluorescent GRCD-RGDS-FITC peptide as described by us beforehand27.
Drug screening
A hydrogel precursor resolution containing 20% w/v PEGDA, 0.1% v/v Irgacure, and 106 cells/mL (DiOC-stained U87 cells) was injected and polymerized within the microfluidic machine cell tradition microwells as described above to organize the non-degradable, non-adhesive PEGDA hydrogels. Instantly after gelation, RPMI media supplemented with 10% FBS and 1% P/S was perfused via the machine for 48 h. At 48 h the perfusion media was changed with supplemented media containing 0.2 μg/mL PI (staining nuclei of lifeless cells) in each inlets and a couple of mM TMZ or 10 µM BCNU in a single inlet and perfused via the machine for 48 h. Equally, degradable, adhesive 4-arm PEG-Ac hydrogels containing 106 cells/mL of DiOC-stained U87 cells, have been ready as described above, cultured for 48 h and uncovered to TMZ for 48 h. Encapsulated cells have been imaged utilizing Axiovert 200 M inverted fluorescence microscope at × 10 magnification. Cell viability was calculated as:
$$Cell ,viability left(%proper)= frac{{N}_{L}}{{N}_{L}+{N}_{D}}occasions 100,$$
(5)
the place NL represents the variety of reside cells and ND represents the variety of lifeless cells.
Fluorescence correlation spectroscopy
Fluorescence correlation spectroscopy (FCS; Zeiss LSM 510 Fluorescence Microscope, Zeiss, Germany) was used to measure in situ diffusivity of a mannequin fluorophore (Atto 655) in a 20% w/v PEGDA hydrogel slab. Hydrogels have been fashioned as described above, with the modification that Atto 655 (0.5 nM) was added to the hydrogel precursor options. For FCS measurements, hydrogels (150 µL) have been ready in an 8 chamber coverglass with #1 German borosilicate backside. Hydrogels have been then soaked in a single day in DMEM medium with out phenol pink and containing 0.5 nM Atto 655 to keep away from concentration-driven diffusion of Atto 655 from the hydrogel into the encompassing media. Atto 655 (0.5 nM) in deionized water was additionally used to calibrate the confocal quantity of the FCS instrument. A 633 nm ps pulsed laser was used for six measurements of 300 s for every pattern location. An autocorrelation perform G(τ) was obtained for every measurement:
$$Gleft(tau proper)=left[frac{1}{N}frac{1}{left[1+left(frac{tau }{{tau }_{D}}right)right]}frac{1}{{left[1+pleft(frac{tau }{{tau }_{D}}right)right]}^{0.5}}proper]left[1+frac{T}{1-T}{e}^{frac{-tau }{{tau }_{T}}}right],$$
(6)
the place N is the variety of fluorescent particles, p = ro/zo is an instrumental fixed, ro is the radius and zo is the axial size of the targeted laser beam spot, τd is the solute diffusion time, T is triplet state amplitude, and τT is the triplet lifetime. The autocorrelation perform was match utilizing a Triplet mannequin to account for the attainable excitation of molecular triplet states at larger laser intensities. Lastly, the autocorrelation perform was normalized as follows:
$$Normalized ,Gleft(tau proper)=G({tau }_{D})/G({tau }_{0}),$$
(7)
the place G(τD) is the worth of Eq. (6) at every time level and G(τ0) is the worth of Eq. (6) on the preliminary time level. The efficient tracer diffusion coefficient for every protein in resolution was calculated from τD as:
$${D}_{FCS}={left({r}_{0}proper)}^{2}/4{tau }_{D}.$$
(8)
Assuming the diffusivities of TMZ and Atto 655 have been related on account of their comparatively related molecular weights (194 g/mol and 887 g/mol, respectively), we then modeled the anticipated TMZ focus within the gels as a perform of gel depth and time. TMZ focus was calculated through Fick’s second regulation of diffusion for 1-D geometry (a skinny slab) with the next boundary situations: c(x = 0) = 2 mM TMZ and c(x = ∞) = 0 mM TMZ at t = 0 h, the place c is the TMZ focus, x is the gel location and t is time:
$$cleft(x,tright)=2 {textual content{mM}}occasions [1-text{erf}left(x/2sqrt(Dt)right)].$$
(9)
Statistical evaluation
All information are introduced as imply values (± SD) decided from 3 to 4 unbiased experiments. Statistical evaluation for focus gradient evaluation and loading reproducibility was carried out utilizing one-way ANOVA with Holm–Sidak or Dunn’s a number of comparability exams utilizing GraphPad Prism 6®. Statistical evaluation for the slope of the depth profile vs volumetric movement charge was carried out utilizing linear-regression evaluation utilizing GraphPad Prism 6®. A p < 0.05 was thought-about statistically important.
Source link